前言
- 本篇主要介绍如何读取edf脑电数据
一、什么是EDF?
-
EDF:
全称是 European Data Format(欧洲数据格式),是一种标准文件格式,用于交换和存储医疗时间序列,其能够存储多通道的数据,允许每个信号拥有不同的采样频率。所以,EDF 也是多导睡眠图(PSG)录音的流行格式。 - EDF的扩展名为EDF +,与EDF基本兼容:所有现有的EDF阅读器也显示EDF +信号;
-
EDF +:
可存储任何医疗记录
,如肌电图,诱发电位,心电图,以及自动和手动分析结果,如δ图,QRS参数和睡眠阶段。
脑电其它常用格式:
心理学MATLAB初学者教程 – 脑电数据读取
二、处理EDF文件
1. 软件读取EDF
常见的读取 EDF 文件的软件有以下几种:
使用EDFbrowser示例(效果如下图所示,还能像视频一样播放):

2. Python 读写EDF
2.1 使用MNE
-
内容详见:
MNE-python读取.edf文件
2.2 使用Pyedflib
-
源码:
官网下载示例
-
解析:
- 读取文件示例:
import numpy as np
import pyedflib
def read_edf(file):
f = pyedflib.EdfReader(file)
print("\nlibrary version: %s" % pyedflib.version.version)
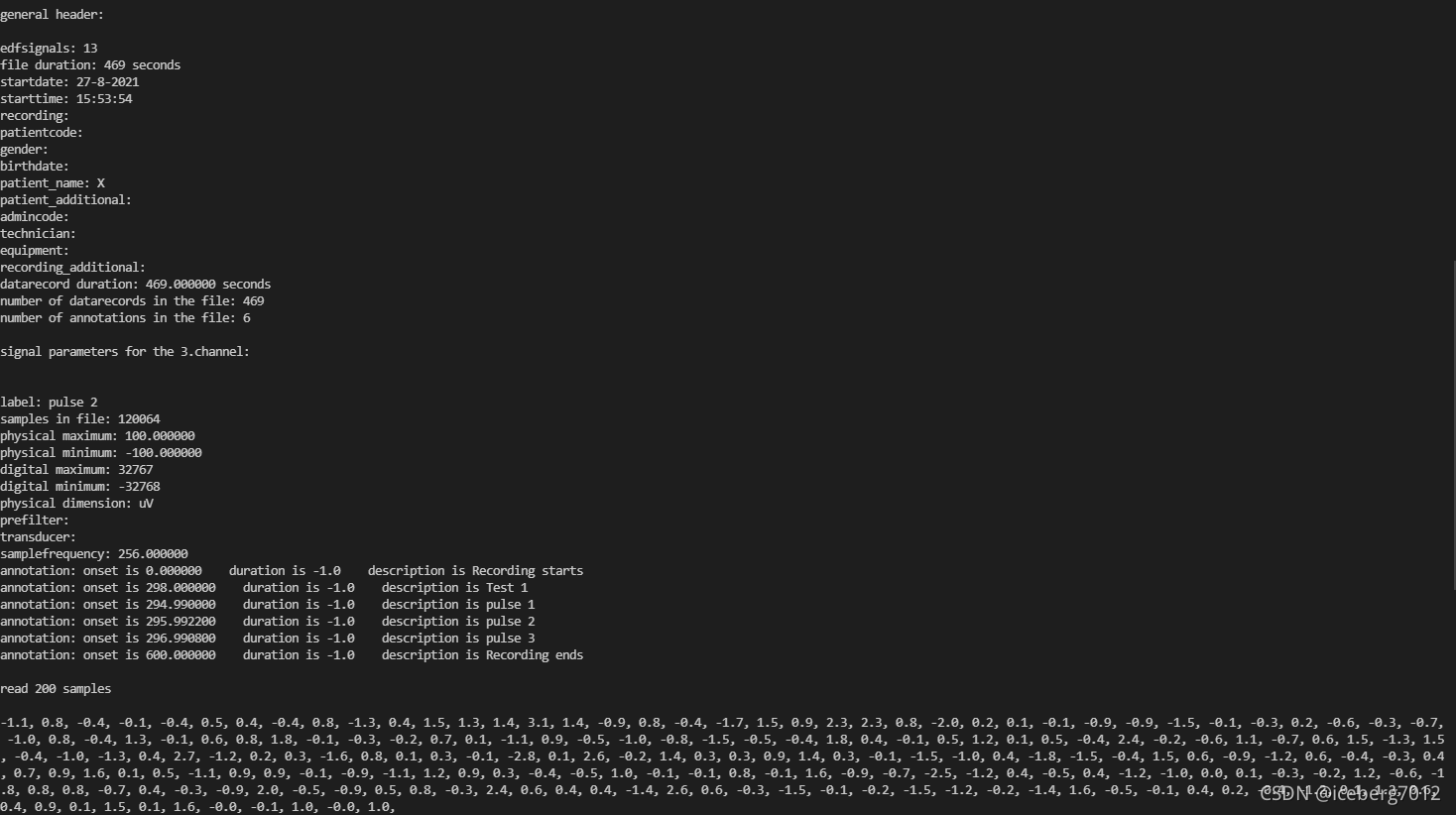
print("\ngeneral header:\n")
print("edfsignals: %i" % f.signals_in_file)
print("file duration: %i seconds" % f.file_duration)
print("startdate: %i-%i-%i" %
(f.getStartdatetime().day, f.getStartdatetime().month,
f.getStartdatetime().year))
print("starttime: %i:%02i:%02i" %
(f.getStartdatetime().hour, f.getStartdatetime().minute,
f.getStartdatetime().second))
print("recording: %s" % f.getPatientAdditional())
print("patientcode: %s" % f.getPatientCode())
print("gender: %s" % f.getGender())
print("birthdate: %s" % f.getBirthdate())
print("patient_name: %s" % f.getPatientName().encode('ascii').decode('unicode_escape'))
print("patient_additional: %s" % f.getPatientAdditional())
print("admincode: %s" % f.getAdmincode())
print("technician: %s" % f.getTechnician())
print("equipment: %s" % f.getEquipment())
print("recording_additional: %s" % f.getRecordingAdditional())
print("datarecord duration: %f seconds" % f.getFileDuration())
print("number of datarecords in the file: %i" % f.datarecords_in_file)
print("number of annotations in the file: %i" % f.annotations_in_file)
channel = 3
print("\nsignal parameters for the %d.channel:\n\n" % channel)
print("label: %s" % f.getLabel(channel))
print("samples in file: %i" % f.getNSamples()[channel])
# print("samples in datarecord: %i" % f.get
print("physical maximum: %f" % f.getPhysicalMaximum(channel))
print("physical minimum: %f" % f.getPhysicalMinimum(channel))
print("digital maximum: %i" % f.getDigitalMaximum(channel))
print("digital minimum: %i" % f.getDigitalMinimum(channel))
print("physical dimension: %s" % f.getPhysicalDimension(channel))
print("prefilter: %s" % f.getPrefilter(channel))
print("transducer: %s" % f.getTransducer(channel))
print("samplefrequency: %f" % f.getSampleFrequency(channel))
annotations = f.readAnnotations()
for n in np.arange(f.annotations_in_file):
print(
"annotation: onset is %f duration is %s description is %s" %
(annotations[0][n], annotations[1][n], annotations[2][n]))
buf = f.readSignal(channel)
n = 200
print("\nread %i samples\n" % n)
result = ""
for i in np.arange(n):
result += ("%.1f, " % buf[i])
print(result)
f.close()
del f
if __name__ == "__main__":
demofilepath = 'demo_write.edf'
read_edf(demofilepath)
运行结果:
- 写入文件示例:(创建名为demo_write的EDF文件,另:可能是版本的关系,源码中的sample_frequency需修改为sample_rate)
import numpy as np
import pyedflib
# signal label/waveform amplitude f sf
# ---------------------------------------------------
# 1 squarewave 100 uV 0.1Hz 200 Hz
# 2 ramp 100 uV 1 Hz 200 Hz
# 3 pulse 1 100 uV 1 Hz 200 Hz
# 4 pulse 2 100 uV 1 Hz 256 Hz
# 5 pulse 3 100 uV 1 Hz 217 Hz
# 6 noise 100 uV - Hz 200 Hz
# 7 sine 1 Hz 100 uV 1 Hz 200 Hz
# 8 sine 8 Hz 100 uV 8 Hz 200 Hz
# 9 sine 8.1777 Hz 100 uV 8.25 Hz 200 Hz
# 10 sine 8.5 Hz 100 uV 8.5Hz 200 Hz
# 11 sine 15 Hz 100 uV 15 Hz 200 Hz
# 12 sine 17 Hz 100 uV 17 Hz 200 Hz
# 13 sine 50 Hz 100 uV 50 Hz 200 Hz
def write_edf(file, channel):
file_duration = 600
f = pyedflib.EdfWriter(file,
channel,
file_type=pyedflib.FILETYPE_EDFPLUS)
channel_info = []
data_list = []
ch_dict = {
'label': 'squarewave',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'ramp',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'pulse 1',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'pulse 2',
'dimension': 'uV',
'sample_rate': 256,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'pulse 3',
'dimension': 'uV',
'sample_rate': 217,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'noise',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 1 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 8 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 8.1777 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 8.5 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 15 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 17 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
ch_dict = {
'label': 'sine 50 Hz',
'dimension': 'uV',
'sample_rate': 200,
'physical_max': 100,
'physical_min': -100,
'digital_max': 32767,
'digital_min': -32768,
'transducer': '',
'prefilter': ''
}
channel_info.append(ch_dict)
data_list.append(np.random.normal(size=file_duration * 200))
f.setSignalHeaders(channel_info)
f.writeSamples(data_list)
f.writeAnnotation(0, -1, "Recording starts")
f.writeAnnotation(298, -1, "Test 1")
f.writeAnnotation(294.99, -1, "pulse 1")
f.writeAnnotation(295.9921875, -1, "pulse 2")
f.writeAnnotation(296.99078341013825, -1, "pulse 3")
f.writeAnnotation(600, -1, "Recording ends")
f.close()
del f
if __name__ == "__main__":
demofilepath = 'demo_write.edf'
write_edf(demofilepath, 13) # 创建写edf样本
- 绘图示例:
# based on multilineplot example in matplotlib with MRI data (I think)
# uses line collections (might actually be from pbrain example)
# clm
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
import pyedflib
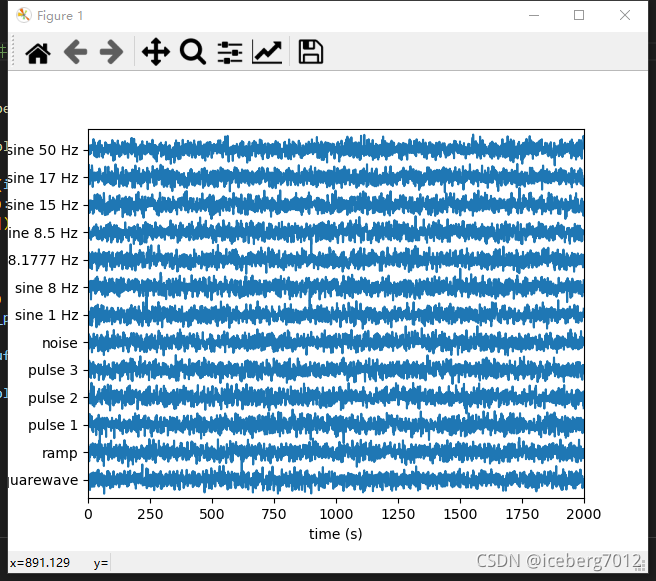
def stackplot(marray, seconds=None, start_time=None, ylabels=None, ax=None):
"""
will plot a stack of traces one above the other assuming
marray.shape = numRows, numSamples
"""
tarray = np.transpose(marray)
stackplot_t(tarray,
seconds=seconds,
start_time=start_time,
ylabels=ylabels,
ax=ax)
plt.show()
def stackplot_t(tarray, seconds=None, start_time=None, ylabels=None, ax=None):
"""
will plot a stack of traces one above the other assuming
tarray.shape = numSamples, numRows
"""
data = tarray
numSamples, numRows = tarray.shape
# data = np.random.randn(numSamples,numRows) # test data
# data.shape = numSamples, numRows
if seconds:
t = seconds * np.arange(numSamples, dtype=float) / numSamples
# import pdb
# pdb.set_trace()
if start_time:
t = t + start_time
xlm = (start_time, start_time + seconds)
else:
xlm = (0, seconds)
else:
t = np.arange(numSamples, dtype=float)
xlm = (0, numSamples)
ticklocs = []
if ax is None:
ax = plt.subplot(111)
plt.xlim(*xlm)
# xticks(np.linspace(xlm, 10))
dmin = data.min()
dmax = data.max()
dr = (dmax - dmin) * 0.7 # Crowd them a bit.
y0 = dmin
y1 = (numRows - 1) * dr + dmax
plt.ylim(y0, y1)
segs = []
for i in range(numRows):
segs.append(np.hstack((t[:, np.newaxis], data[:, i, np.newaxis])))
# print "segs[-1].shape:", segs[-1].shape
ticklocs.append(i * dr)
offsets = np.zeros((numRows, 2), dtype=float)
offsets[:, 1] = ticklocs
lines = LineCollection(
segs,
offsets=offsets,
transOffset=None,
)
ax.add_collection(lines)
# set the yticks to use axes coords on the y axis
ax.set_yticks(ticklocs)
# ax.set_yticklabels(['PG3', 'PG5', 'PG7', 'PG9'])
# if not plt.ylabels:
plt.ylabels = ["%d" % ii for ii in range(numRows)]
ax.set_yticklabels(ylabels)
plt.xlabel('time (s)')
def test_stacklineplot():
numSamples, numRows = 800, 5
data = np.random.randn(numRows, numSamples) # test data
stackplot(data, 10.0)
if __name__ == "__main__":
file = 'demo_write.edf' # 文件路径
f = pyedflib.EdfReader(file)
n = f.signals_in_file
signal_labels = f.getSignalLabels()
n_min = f.getNSamples()[0]
sigbufs = [np.zeros(f.getNSamples()[i]) for i in np.arange(n)]
for i in np.arange(n):
sigbufs[i] = f.readSignal(i)
if n_min < len(sigbufs[i]):
n_min = len(sigbufs[i])
f._close()
del f
n_plot = np.min((n_min, 2000))
sigbufs_plot = np.zeros((n, n_plot))
for i in np.arange(n):
sigbufs_plot[i, :] = sigbufs[i][:n_plot]
stackplot(sigbufs_plot[:, :n_plot], ylabels=signal_labels)
运行结果:
3. C/C++ 读写EDF
本文参考来源:
版权声明:本文为iceberg7012原创文章,遵循 CC 4.0 BY-SA 版权协议,转载请附上原文出处链接和本声明。