import numpy as np
import pandas as pd
import lightgbm as lgb
import sklearn
import scipy
import gc
import missingno
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import classification_report,accuracy_score
from pyod.models.iforest import IForest
RANDOM_SEED = 42
LABELS = ["Normal", "Fraud"]
import plotly.graph_objs as go
import plotly
import plotly.figure_factory as ff
from plotly.offline import init_notebook_mode, iplot
import random
import os
from sklearn.preprocessing import *
from sklearn.model_selection import train_test_split
from sklearn.model_selection import StratifiedKFold
from sklearn.linear_model import *
from sklearn.metrics import *
def SeedEverything(seed):
random.seed(seed)
os.environ['PYTHONHASHSEED']=str(seed)
np.random.seed(seed)
return
from sklearn.linear_model import LogisticRegression
def reduce_mem_usage(df):
start_mem = df.memory_usage().sum() / 1024**2
print('Memory usage of dataframe is {:.2f} MB'.format(start_mem))
for col in df.columns:
col_type = df[col].dtype
if col_type != object:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == 'int':
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:
df[col] = df[col].astype(np.float16)
elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
else:
df[col] = df[col].astype('category')
end_mem = df.memory_usage().sum() / 1024**2
print('Memory usage after optimization is: {:.2f} MB'.format(end_mem))
print('Decreased by {:.1f}%'.format(100 * (start_mem - end_mem) / start_mem))
return df
data = pd.read_csv('creditcard_data.csv',sep=',')
print(data.columns)
#Index(['Time', 'V1', 'V2', 'V3', 'V4', 'V5', 'V6', 'V7', 'V8', 'V9', 'V10',
# 'V11', 'V12', 'V13', 'V14', 'V15', 'V16', 'V17', 'V18', 'V19', 'V20',
# 'V21', 'V22', 'V23', 'V24', 'V25', 'V26', 'V27', 'V28', 'Amount',
# 'Class'],
# dtype='object')
data
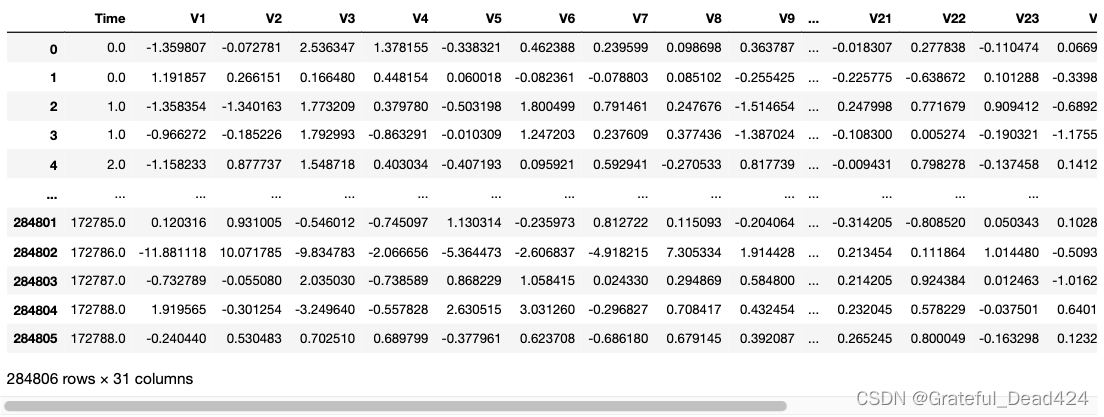
很遗憾,大部分特征都经过加密处理,只有time和amount是未加密的数据,class表示样本的类别,0表示正常样本,1表示异常样本。
missingno.matrix(data)# 查看缺失值情况
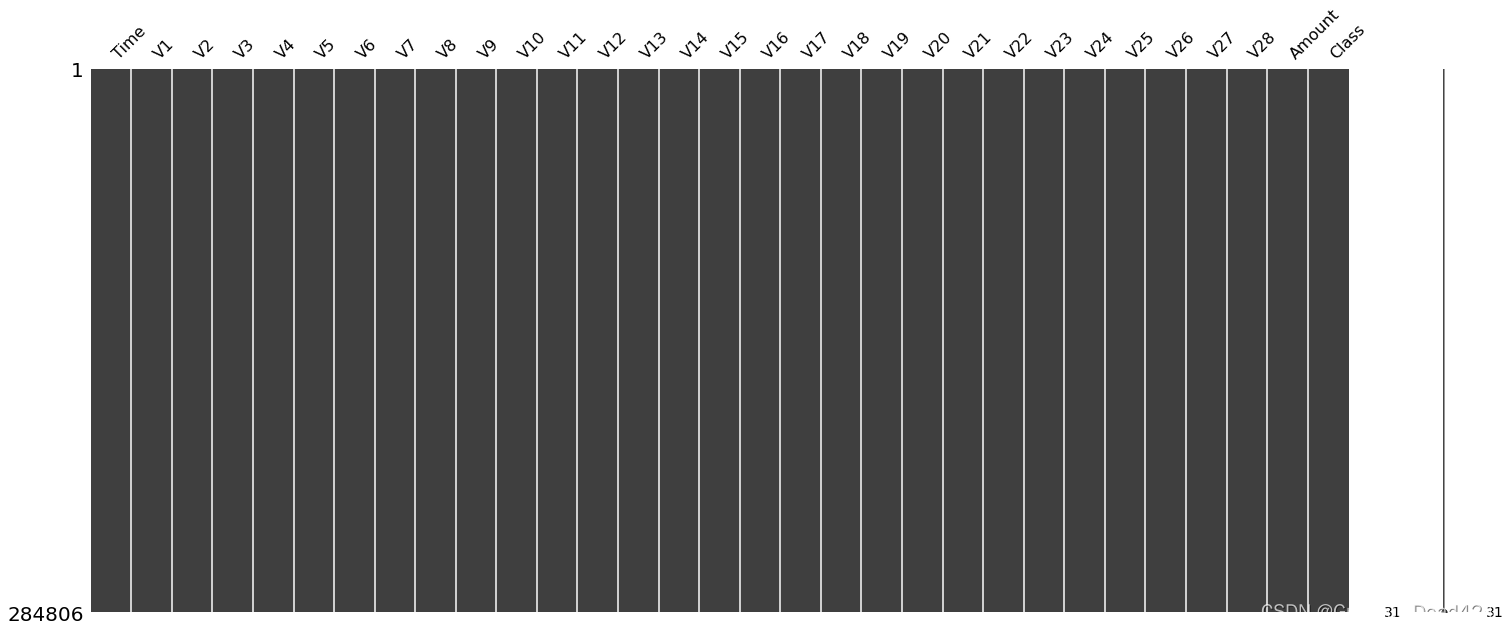
数据质量很高,没有缺失值并且全是连续类型的特征,这对于很多无监督算法来说都是很便利的。
data['Class'].value_counts()
#0 284314
#1 492
#Name: Class, dtype: int64
异常样本占比:0.001727485630620034,大概是0.17%左右,极度不均衡的分类样本了。
我们把time转化为hour然后对所有数据做个标准化之后开始模型训练:
data['Hour'] =data["Time"].apply(lambda x : divmod(x, 3600)[0])
X=data.drop(['Time','Class'],axis=1)
y=data.Class
sd=StandardScaler()
column=X.columns
X[column]=sd.fit_transform(X[column])
1、把异常检测算法转化为有监督的分类问题:
大部分情况下,有监督学习的模型要比无监督强大很多,所以我们首先尝试一下使用有监督+不均衡学习的方案来处理当前问题。
SeedEverything(123) #严格控制随机性
X=reduce_mem_usage(X) #降低原始数据的内存占用
X,X_test,y,y_test=train_test_split(X,y,stratify=y,test_size=0.2) #训练集交叉验证,开发集测试泛化性能
#Memory usage of dataframe is 65.19 MB
#Memory usage after optimization is: 16.30 MB
#Decreased by 75.0%
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_preds = np.zeros(X_test.shape[0])
y_oof = np.zeros(X.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
clf = LogisticRegression(random_state=123,n_jobs=12)
clf.fit(X_train,y_train)
y_pred_valid = clf.predict_proba(X_valid)[:,1]
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict_proba(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
#Fold 1 | AUC: 0.9787506504420224
#Fold 2 | AUC: 0.979044775454892
#Fold 3 | AUC: 0.9840376658105412
#Fold 4 | AUC: 0.9780686254594833
#Fold 5 | AUC: 0.9706782555760354
#
#Mean AUC = 0.9781159945485949
#Out of folds AUC = 0.9780967535677503
#test datasets AUC = 0.960284222721165
这个数据的可分性太简单了。逻辑回归都可以得到很不错的结果
再一次验证了,很多时候不平衡问题对分类模型带来的影响并不是不平衡本身而是数据本身复杂的分类难度,难分类的数据即使是平衡也很难训练出好的模型,所以面对不平衡问题优先考虑的 是样本工程和 特征工程。
params={'num_leaves':491,
'min_child_weight': 0.03454472573214212,
'feature_fraction': 0.3797454081646243,
'bagging_fraction': 0.4181193142567742,
'min_data_in_leaf': 106,
'objective': 'binary',
'max_depth': -1,
'learning_rate': 0.006883242363721497,
"boosting_type": "gbdt",
"bagging_seed": 11,
"metric": 'auc',
"verbosity": -1,
'reg_alpha': 0.3899927210061127,
'reg_lambda': 0.6485237330340494,
'random_state': 47,
}
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
feature_importances = pd.DataFrame()
feature_importances['feature'] = columns
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
dtrain = lgb.Dataset(X_train, label=y_train)
dvalid = lgb.Dataset(X_valid, label=y_valid)
clf = lgb.train(params, dtrain, 10000, valid_sets = [dtrain, dvalid], verbose_eval=200, early_stopping_rounds=200)
feature_importances[f'fold_{fold_n + 1}'] = clf.feature_importance()
y_pred_valid = clf.predict(X_valid)
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict(X_test) / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
# Training until validation scores don't improve for 200 rounds
# [200] training's auc: 0.999659 valid_1's auc: 0.973216
# [400] training's auc: 0.99996 valid_1's auc: 0.972691
# Early stopping, best iteration is:
# [259] training's auc: 0.999825 valid_1's auc: 0.974633
# Fold 1 | AUC: 0.9746331785258131
# Training until validation scores don't improve for 200 rounds
# [200] training's auc: 0.999856 valid_1's auc: 0.97012
# [400] training's auc: 0.999977 valid_1's auc: 0.974295
# [600] training's auc: 0.999996 valid_1's auc: 0.975384
# Early stopping, best iteration is:
# [459] training's auc: 0.999986 valid_1's auc: 0.97573
# Fold 2 | AUC: 0.9757300950827975
# Training until validation scores don't improve for 200 rounds
# [200] training's auc: 0.999165 valid_1's auc: 0.98765
# [400] training's auc: 0.999934 valid_1's auc: 0.991137
# [600] training's auc: 0.999993 valid_1's auc: 0.991546
# [800] training's auc: 0.999999 valid_1's auc: 0.992762
# [1000] training's auc: 1 valid_1's auc: 0.994013
# [1200] training's auc: 1 valid_1's auc: 0.994248
# [1400] training's auc: 1 valid_1's auc: 0.994315
# [1600] training's auc: 1 valid_1's auc: 0.994258
# Early stopping, best iteration is:
# [1510] training's auc: 1 valid_1's auc: 0.994366
# Fold 3 | AUC: 0.994366267728893
# Training until validation scores don't improve for 200 rounds
# [200] training's auc: 0.999523 valid_1's auc: 0.976617
# Early stopping, best iteration is:
# [40] training's auc: 0.996496 valid_1's auc: 0.982966
# Fold 4 | AUC: 0.9829664886704828
# Training until validation scores don't improve for 200 rounds
# [200] training's auc: 0.999774 valid_1's auc: 0.985556
# [400] training's auc: 0.99997 valid_1's auc: 0.987184
# [600] training's auc: 0.999994 valid_1's auc: 0.987817
# Early stopping, best iteration is:
# [517] training's auc: 0.999989 valid_1's auc: 0.987999
# Fold 5 | AUC: 0.9879993348777697
# Mean AUC = 0.9831390729771512
# Out of folds AUC = 0.9673819593306054
# test datasets AUC = 0.9812737946895134
lgb的性能一般情况下是要好过lr的没什么问题
feature_importances['average'] = feature_importances[[f'fold_{fold_n + 1}' for fold_n in range(folds.n_splits)]].mean(axis=1)
feature_importances.to_csv('feature_importances.csv')
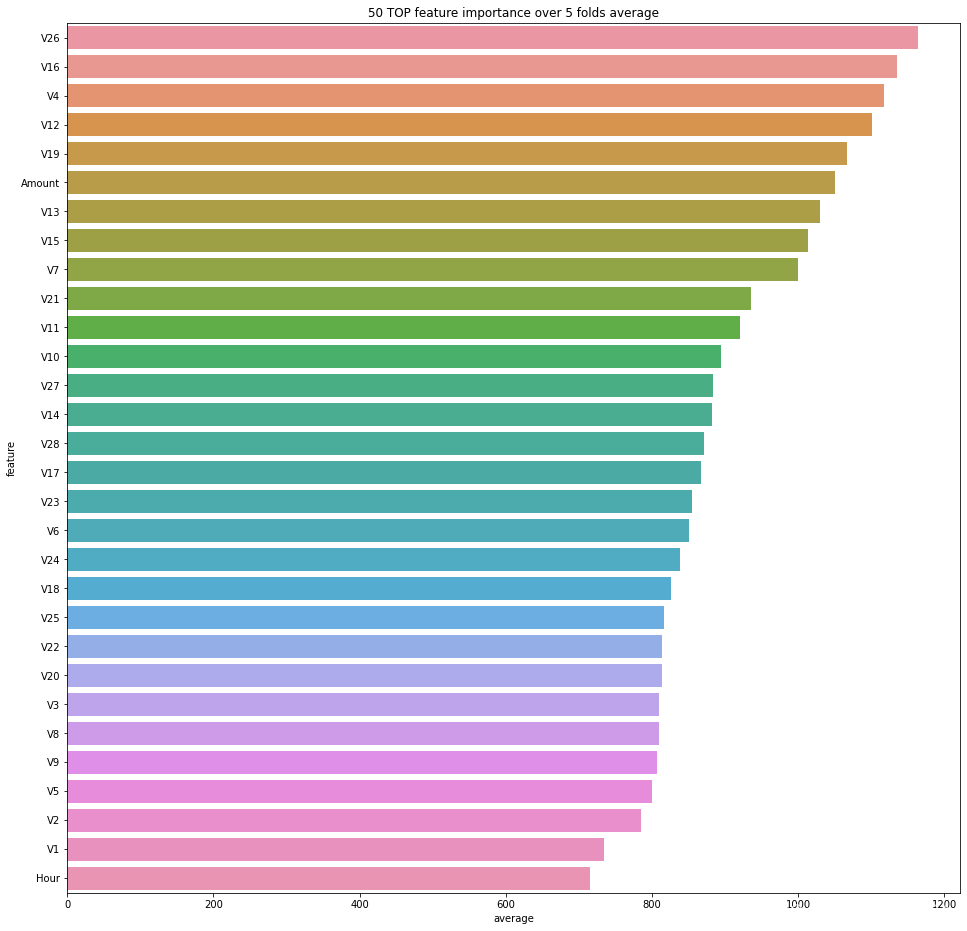
plt.figure(figsize=(16, 16))
sns.barplot(data=feature_importances.sort_values(by='average', ascending=False).head(50), x='average', y='feature');
plt.title('50 TOP feature importance over {} folds average'.format(folds.n_splits));
使用专门的异常检测算法:
因为聚类系列本身的算法是比较多的,所以这里暂时先讨论一些非聚类的异常检测算法。
自编码器
自编码器进行异常检测的思路很简单,就是重构误差作为异常程度的衡量标准,用自编码器对原始数据进行编码与解码然后通过mse、欧氏距离等度量方式来衡量重构误差,重构误差越大越容易是异常样本。和聚类不一样,自编码器不会受到维度诅咒的影响,因为其本身就是可以类似pca的对原始数据进行压缩,将原始数据用更低维度的特征来表示进行表征学习。算法的基本假设就是大部分正常样本服从相似的分布映射的过程中在高低维的位置接近,而异常样本服从不同的分布,重构的过程中并不满足大部分正常样本下训练出来的nn所表示的映射关系从而造成较大的重构误差。
当然,自编码器本身的知识体量也不小包括了稀疏自编码、降噪自编码器、vae等
autoencoder严格上来说更接近novelty detection,模型学习的是原始正常数据的某种映射关系,和oneclasssvm一样,如果我们初始的数据仅仅占了全部数据的一部分,那么后期预测就很容易把未训练过的新的正常的样本预测为异常样本了。
例如这种数据分布,假设我们一开始使用的是左下角的样本训练,则测试的时候右上角的正常样本基本全部都会被预测为异常样本
from tensorflow.keras.layers import *
from tensorflow.keras import layers
from tensorflow.keras import Model
from pyod.utils.stat_models import pairwise_distances_no_broadcast
from tensorflow.keras.callbacks import *
import tensorflow as tf
import tensorflow_addons as tfa
es=tf.keras.callbacks.EarlyStopping(
monitor='val_mae', min_delta=0, patience=10, verbose=1,
mode='min', baseline=None, restore_best_weights=True
)
rp=tf.keras.callbacks.ReduceLROnPlateau(
monitor="val_mae",
factor=0.5,
patience=5,
verbose=1,
mode="min",
min_delta=0.0001,
cooldown=0,
min_lr=0.001,
)
def create_model():
encoding_dim = 128
batch_size = 256
input_dim=30
input_layer = Input(shape=(input_dim, ))
encoder = Dense(encoding_dim,kernel_regularizer='l2')(input_layer)
encoder=tfa.activations.mish(encoder)
encoder = Dense(int(encoding_dim / 2),kernel_regularizer='l2')(encoder)
encoder=tfa.activations.mish(encoder)
encoder = Dense(int(encoding_dim / 4),kernel_regularizer='l2')(encoder)
decoder=tfa.activations.mish(encoder)
decoder = Dense(encoding_dim / 4,kernel_regularizer='l2')(decoder)
decoder=tfa.activations.mish(decoder)
decoder = Dense(encoding_dim / 2,kernel_regularizer='l2')(decoder)
decoder=tfa.activations.mish(decoder)
decoder = Dense(encoding_dim ,kernel_regularizer='l2')(decoder)
decoder=tfa.activations.mish(decoder)
decoder = Dense(input_dim,kernel_regularizer='l2')(decoder)
autoencoder = Model(inputs=input_layer, outputs=decoder)
autoencoder.compile(optimizer=tf.keras.optimizers.Adam(0.005),
loss='mean_absolute_error',
metrics=['mae'])
return autoencoder
num_epoch = 1000
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
autoencoder=create_model()
autoencoder.fit(X_train, X_train,validation_data=(X_valid,y_valid),callbacks=[es,rp],
epochs=num_epoch,
batch_size=512,
shuffle=True,
verbose=1,)
y_pred_valid = autoencoder.predict(X_valid)
y_oof[valid_index] = pairwise_distances_no_broadcast(X_valid.astype('float').values,y_pred_valid.astype('float'))
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_oof[valid_index])}")
score += roc_auc_score(y_valid, y_oof[valid_index]) / NFOLDS
y_pred=pairwise_distances_no_broadcast(X_test.astype('float').values,autoencoder.predict(X_test).astype('float'))
y_preds += y_pred/ NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
最终我们使用欧式距离来作为异常程度的衡量,整体的效果不错,达到了0.95+左右的auc,对于一个无监督的算法而言已经是不错的结果了
孤立森林
直接看一下pyod中的iforest参数吧,与算法原理有关的就n_estimators,max_samples和max_feaures以及bootstrap.
需要注意的是,iforest会根据样本的数目来自动设置树的深度:
因为一般我们只关心异常样本,也就是切分路径短的样本,而正常样本的深度是n还是2n其实无所谓,这样的深度设计在很大程度上降低了模型的训练时间提高了训练效率。不过这里为什么用log2倒是没有明确的说明。所以在iforest的算法设计上树的深度没有作为超参数存在
局限性:
(1)如果训练样本中异常样本的比例比较高,违背了先前提到的异常检测的基本假设,可能最终的效果会受影响,这类问题在欺诈团伙作案的应用场景上比较常见,欺诈分子的样本不小,比如;
(2)异常检测跟具体的应用场景紧密相关,算法检测出的“异常”不一定是我们实际想要的。比如,在识别虚假交易时,异常的交易未必就是虚假的交易。所以,在特征选择时,可能需要过滤不太相关的特征,以免识别出一些不太相关的“异常”,这是大部分异常检测算法的通病了,包括聚类,有一些特征和区分正常和异常样本根本没有帮助,但是也会引入计算导致影响最终结果,这涉及到异常检测的特征选择了,后续在异常检测专栏总结。
经验值 maxsample=256 n_estiamtros=100
接下来我们就用iforest来测试下效果吧
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
clf=IForest(n_estimators=100, max_samples='auto', contamination=0.1, \
max_features=1.0, bootstrap=False, n_jobs=12, behaviour='old', random_state=123, verbose=1)
clf.fit(X_train)
y_pred_valid = clf.predict_proba(X_valid)[:,1]
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict_proba(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
#Fold 1 | AUC: 0.9404957550831871
#Fold 2 | AUC: 0.937875343308169
#Fold 3 | AUC: 0.9529438936363813
#Fold 4 | AUC: 0.9516246441699525
#Fold 5 | AUC: 0.9472558071370998
#Mean AUC = 0.9460390886669579
#Out of folds AUC = 0.9457023075300758
#test datasets AUC = 0.9478101348868191
可以看到孤立森林的效果也不错,仅仅使用简单的参数,相对而言已经比较接近有监督学习算法的精度了,相对于自编码器而言,开箱即用性强,自编码器我花了不少时间设计网络结构才能达到不错的效果
from pyod.models.knn import KNN
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
clf=KNN(contamination=0.1, n_neighbors=5, method='largest', radius=1.0, \
algorithm='auto', leaf_size=30, metric='minkowski', p=2,n_jobs=12)
clf.fit(X_train)
y_pred_valid = clf.predict_proba(X_valid)[:,1]
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict_proba(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
#Fold 1 | AUC: 0.9484273911918325
#Fold 2 | AUC: 0.9307021156409393
#Fold 3 | AUC: 0.9555083743540798
knn整体效果还可以,但是knn以及其它的无论是基于密度还是基于距离的算法的致命问题就在于计算太慢,并且内存占用一般都比较高
LOF
基于密度,lof和dbscan以及optimics这类以密度为计算核心的算法类似,都是通过样本在空间中的稀疏程度来定义异常值的
from pyod.models.lof import LOF
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
clf=LOF(n_jobs=12)
clf.fit(X_train)
y_pred_valid = clf.predict_proba(X_valid)[:,1]
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict_proba(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
#Mean AUC = 0.4934845376918889
#Out of folds AUC = 0.49510929495298234
#test datasets AUC = 0.47531327880054663
数据的分布并不符合lof的基本假设,结果比较渣。考虑到可能 是正负样本本身就是两个完整的群体
oneclass svm
最后我们再来看一下oneclasssvm的效果
from pyod.models.ocsvm import OneClassSVM
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=2021,shuffle=True)
columns = X.columns
splits = folds.split(X, y)
y_oof = np.zeros(X.shape[0])
y_preds = np.zeros(X_test.shape[0])
score = 0
for fold_n, (train_index, valid_index) in enumerate(splits):
X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]
y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]
clf=OneClassSVM()
clf.fit(X_train)
y_pred_valid = clf.decision_function(X_valid)[:,1]
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.decision_function(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
gc.collect()
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")